Total Variation Denoising with Constraint (APGM)#
This example demonstrates the solution of the isotropic total variation (TV) denoising problem
where \(R\) is a TV regularizer, \(\iota_C(\cdot)\) is the indicator function of constraint set \(C\), and \(C = \{ \mathbf{x} \, | \, x_i \in [0, 1] \}\), i.e. the set of vectors with components constrained to be in the interval \([0, 1]\). The problem is solved seperately with \(R\) taken as isotropic and anisotropic TV regularization
The solution via APGM is based on the approach in [9], which involves constructing a dual for the constrained denoising problem. The APGM solution minimizes the resulting dual. In this case, switching between the two regularizers corresponds to switching between two different projectors.
[1]:
from typing import Callable, Optional, Union
import jax.numpy as jnp
from xdesign import SiemensStar, discrete_phantom
import scico.numpy as snp
import scico.random
from scico import functional, linop, loss, operator, plot
from scico.numpy import Array, BlockArray
from scico.optimize.pgm import AcceleratedPGM, RobustLineSearchStepSize
from scico.util import device_info
plot.config_notebook_plotting()
Create a ground truth image.
[2]:
N = 256 # image size
phantom = SiemensStar(16)
x_gt = snp.pad(discrete_phantom(phantom, N - 16), 8)
x_gt = x_gt / x_gt.max()
Add noise to create a noisy test image.
[3]:
σ = 0.75 # noise standard deviation
noise, key = scico.random.randn(x_gt.shape, seed=0)
y = x_gt + σ * noise
Define finite difference operator and adjoint.
[4]:
# The append=0 option appends 0 to the input along the axis
# prior to performing the difference to make the results of
# horizontal and vertical finite differences the same shape.
C = linop.FiniteDifference(input_shape=x_gt.shape, append=0)
A = C.adj
Define a zero array as initial estimate.
[5]:
x0 = jnp.zeros(C(y).shape)
Define the dual of the total variation denoising problem.
[6]:
class DualTVLoss(loss.Loss):
def __init__(
self,
y: Union[Array, BlockArray],
A: Optional[Union[Callable, operator.Operator]] = None,
lmbda: float = 0.5,
):
self.functional = functional.SquaredL2Norm()
super().__init__(y=y, A=A, scale=1.0)
self.lmbda = lmbda
def __call__(self, x: Union[Array, BlockArray]) -> float:
xint = self.y - self.lmbda * self.A(x)
return -1.0 * self.functional(xint - jnp.clip(xint, 0.0, 1.0)) + self.functional(xint)
Denoise with isotropic total variation. Define projector for isotropic total variation.
[7]:
# Evaluation of functional set to zero.
class IsoProjector(functional.Functional):
has_eval = True
has_prox = True
def __call__(self, x: Union[Array, BlockArray]) -> float:
return 0.0
def prox(self, v: Array, lam: float, **kwargs) -> Array:
norm_v_ptp = jnp.sqrt(jnp.sum(jnp.abs(v) ** 2, axis=0))
x_out = v / jnp.maximum(jnp.ones(v.shape), norm_v_ptp)
out1 = v[0, :, -1] / jnp.maximum(jnp.ones(v[0, :, -1].shape), jnp.abs(v[0, :, -1]))
x_out = x_out.at[0, :, -1].set(out1)
out2 = v[1, -1, :] / jnp.maximum(jnp.ones(v[1, -1, :].shape), jnp.abs(v[1, -1, :]))
x_out = x_out.at[1, -1, :].set(out2)
return x_out
Use RobustLineSearchStepSize object and set up AcceleratedPGM solver object. Run the solver.
[8]:
reg_weight_iso = 1.4e0
f_iso = DualTVLoss(y=y, A=A, lmbda=reg_weight_iso)
g_iso = IsoProjector()
solver_iso = AcceleratedPGM(
f=f_iso,
g=g_iso,
L0=16.0 * f_iso.lmbda**2,
x0=x0,
maxiter=100,
itstat_options={"display": True, "period": 10},
step_size=RobustLineSearchStepSize(),
)
# Run the solver.
print(f"Solving on {device_info()}\n")
x = solver_iso.solve()
hist_iso = solver_iso.itstat_object.history(transpose=True)
# Project to constraint set.
x_iso = jnp.clip(y - f_iso.lmbda * f_iso.A(x), 0.0, 1.0)
Solving on GPU (NVIDIA GeForce RTX 2080 Ti)
Iter Time Objective L Residual
-----------------------------------------------
0 4.24e+00 3.141e+04 2.822e+01 1.835e+01
10 4.56e+00 1.749e+04 1.968e+01 9.306e+00
20 4.78e+00 1.513e+04 2.745e+01 5.370e+00
30 4.98e+00 1.472e+04 1.914e+01 3.691e+00
40 5.17e+00 1.461e+04 2.670e+01 2.082e+00
50 5.35e+00 1.456e+04 1.862e+01 1.982e+00
60 5.58e+00 1.453e+04 2.597e+01 1.394e+00
70 5.75e+00 1.451e+04 1.811e+01 1.484e+00
80 5.93e+00 1.450e+04 2.526e+01 1.130e+00
90 6.09e+00 1.449e+04 1.761e+01 1.245e+00
99 6.26e+00 1.449e+04 2.729e+01 9.203e-01
Denoise with anisotropic total variation for comparison. Define projector for anisotropic total variation.
[9]:
# Evaluation of functional set to zero.
class AnisoProjector(functional.Functional):
has_eval = True
has_prox = True
def __call__(self, x: Union[Array, BlockArray]) -> float:
return 0.0
def prox(self, v: Array, lam: float, **kwargs) -> Array:
return v / jnp.maximum(jnp.ones(v.shape), jnp.abs(v))
Use RobustLineSearchStepSize object and set up AcceleratedPGM solver object. Weight was tuned to give the same data fidelity as the isotropic case. Run the solver.
[10]:
reg_weight_aniso = 1.2e0
f = DualTVLoss(y=y, A=A, lmbda=reg_weight_aniso)
g = AnisoProjector()
solver = AcceleratedPGM(
f=f,
g=g,
L0=16.0 * f.lmbda**2,
x0=x0,
maxiter=100,
itstat_options={"display": True, "period": 10},
step_size=RobustLineSearchStepSize(),
)
# Run the solver.
print()
x = solver.solve()
# Project to constraint set.
x_aniso = jnp.clip(y - f.lmbda * f.A(x), 0.0, 1.0)
Iter Time Objective L Residual
-----------------------------------------------
0 3.00e-01 3.141e+04 2.074e+01 2.141e+01
10 5.07e-01 1.753e+04 1.446e+01 1.073e+01
20 6.86e-01 1.518e+04 2.017e+01 6.434e+00
30 8.52e-01 1.475e+04 1.406e+01 4.318e+00
40 1.03e+00 1.465e+04 1.962e+01 2.370e+00
50 1.20e+00 1.460e+04 1.368e+01 2.274e+00
60 1.38e+00 1.457e+04 1.908e+01 1.588e+00
70 1.56e+00 1.456e+04 1.330e+01 1.674e+00
80 1.74e+00 1.455e+04 1.856e+01 1.270e+00
90 1.91e+00 1.454e+04 1.294e+01 1.393e+00
99 2.10e+00 1.453e+04 2.005e+01 1.017e+00
Compute the data fidelity.
[11]:
df = hist_iso.Objective[-1]
print(f"\nData fidelity for isotropic TV was {df:.2e}")
hist = solver.itstat_object.history(transpose=True)
df = hist.Objective[-1]
print(f"Data fidelity for anisotropic TV was {df:.2e}")
Data fidelity for isotropic TV was 1.45e+04
Data fidelity for anisotropic TV was 1.45e+04
Plot results.
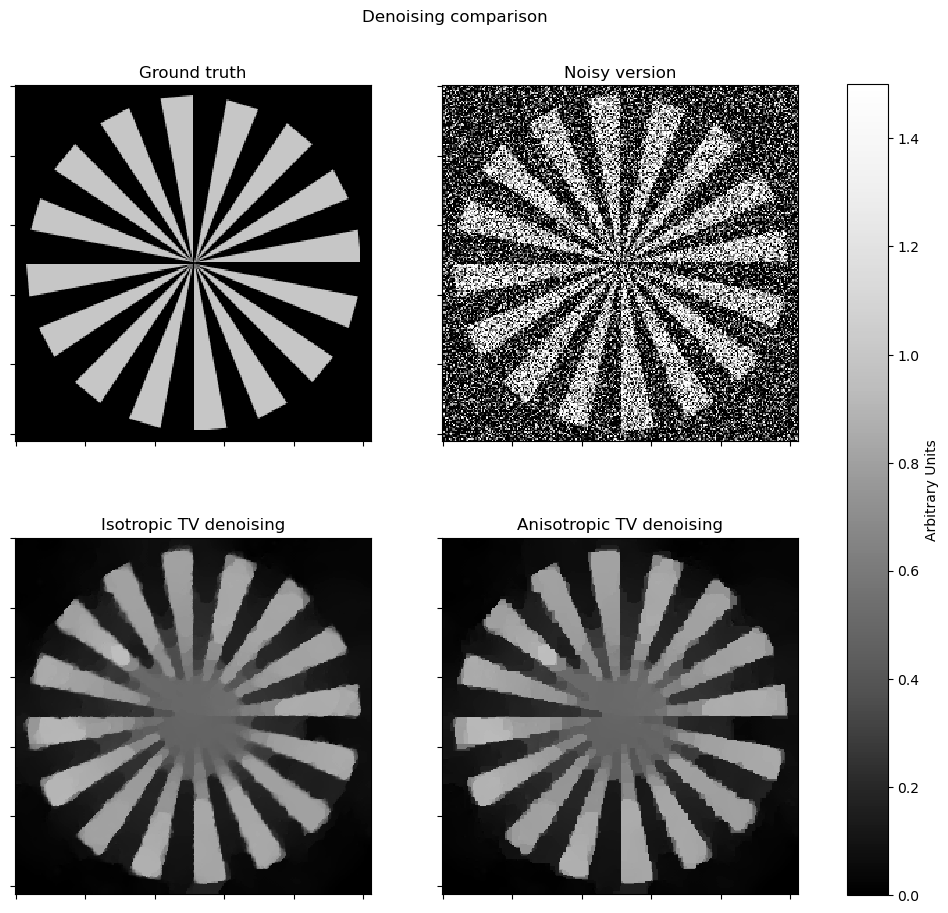
[12]:
plt_args = dict(norm=plot.matplotlib.colors.Normalize(vmin=0, vmax=1.5))
fig, ax = plot.subplots(nrows=2, ncols=2, sharex=True, sharey=True, figsize=(11, 10))
plot.imview(x_gt, title="Ground truth", fig=fig, ax=ax[0, 0], **plt_args)
plot.imview(y, title="Noisy version", fig=fig, ax=ax[0, 1], **plt_args)
plot.imview(x_iso, title="Isotropic TV denoising", fig=fig, ax=ax[1, 0], **plt_args)
plot.imview(x_aniso, title="Anisotropic TV denoising", fig=fig, ax=ax[1, 1], **plt_args)
fig.subplots_adjust(left=0.1, right=0.99, top=0.95, bottom=0.05, wspace=0.2, hspace=0.01)
fig.colorbar(
ax[0, 0].get_images()[0], ax=ax, location="right", shrink=0.9, pad=0.05, label="Arbitrary Units"
)
fig.suptitle("Denoising comparison")
fig.show()
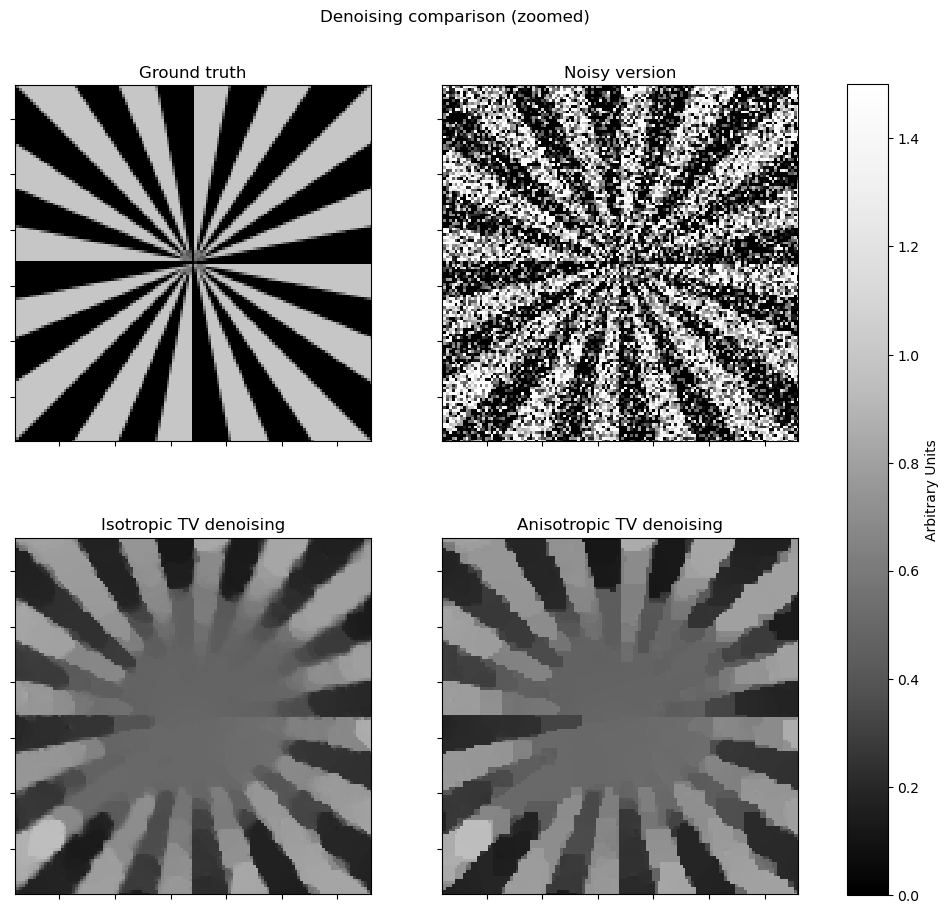
# zoomed version
fig, ax = plot.subplots(nrows=2, ncols=2, sharex=True, sharey=True, figsize=(11, 10))
plot.imview(x_gt, title="Ground truth", fig=fig, ax=ax[0, 0], **plt_args)
plot.imview(y, title="Noisy version", fig=fig, ax=ax[0, 1], **plt_args)
plot.imview(x_iso, title="Isotropic TV denoising", fig=fig, ax=ax[1, 0], **plt_args)
plot.imview(x_aniso, title="Anisotropic TV denoising", fig=fig, ax=ax[1, 1], **plt_args)
ax[0, 0].set_xlim(N // 4, N // 4 + N // 2)
ax[0, 0].set_ylim(N // 4, N // 4 + N // 2)
fig.subplots_adjust(left=0.1, right=0.99, top=0.95, bottom=0.05, wspace=0.2, hspace=0.01)
fig.colorbar(
ax[0, 0].get_images()[0], ax=ax, location="right", shrink=0.9, pad=0.05, label="Arbitrary Units"
)
fig.suptitle("Denoising comparison (zoomed)")
fig.show()