Total Variation Denoising (ADMM)#
This example compares denoising via isotropic and anisotropic total variation (TV) regularization [44] [26]. It solves the denoising problem
\[\mathrm{argmin}_{\mathbf{x}} \; (1/2) \| \mathbf{y} - \mathbf{x}
\|_2^2 + \lambda R(\mathbf{x}) \;,\]
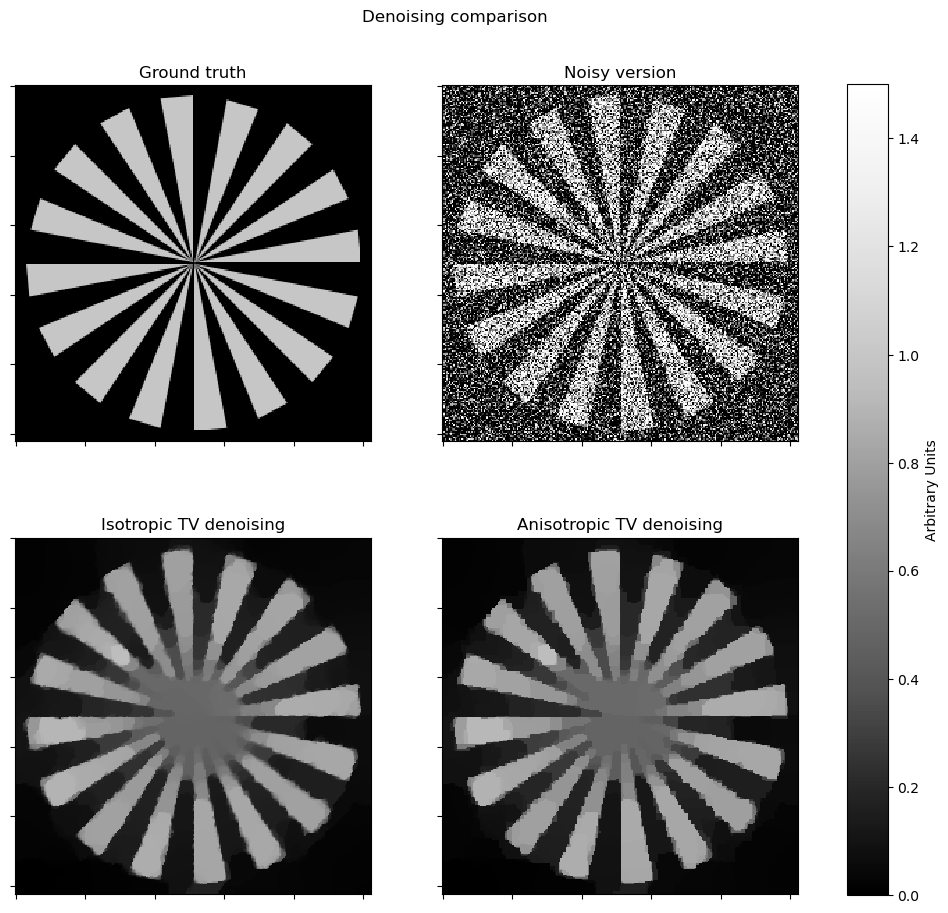
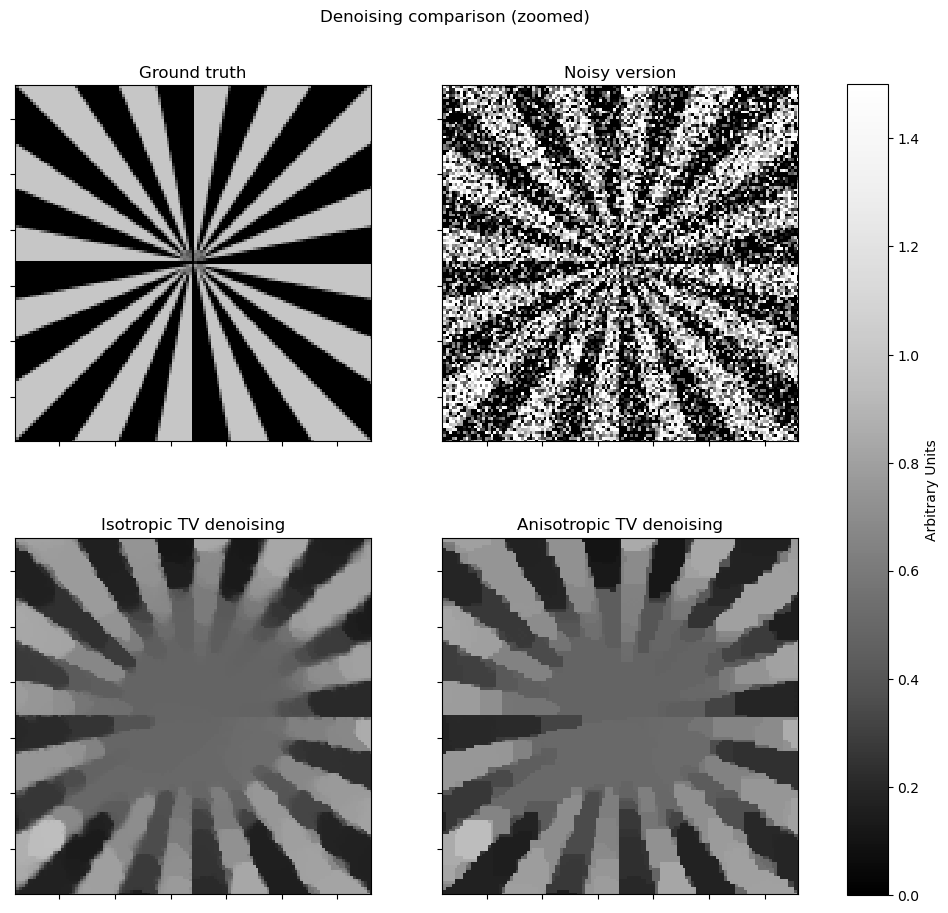
where \(R\) is either the isotropic or anisotropic TV regularizer. In SCICO, switching between these two regularizers is a one-line change: replacing an L1Norm with a L21Norm. Note that the isotropic version exhibits fewer block-like artifacts on edges that are not vertical or horizontal.
[1]:
from xdesign import SiemensStar, discrete_phantom
import scico.numpy as snp
import scico.random
from scico import functional, linop, loss, plot
from scico.optimize.admm import ADMM, LinearSubproblemSolver
from scico.util import device_info
plot.config_notebook_plotting()
Create a ground truth image.
[2]:
N = 256 # image size
phantom = SiemensStar(16)
x_gt = snp.pad(discrete_phantom(phantom, N - 16), 8)
x_gt = x_gt / x_gt.max()
Add noise to create a noisy test image.
[3]:
σ = 0.75 # noise standard deviation
noise, key = scico.random.randn(x_gt.shape, seed=0)
y = x_gt + σ * noise
Denoise with isotropic total variation.
[4]:
λ_iso = 1.4e0
f = loss.SquaredL2Loss(y=y)
g_iso = λ_iso * functional.L21Norm()
# The append=0 option makes the results of horizontal and vertical finite
# differences the same shape, which is required for the L21Norm.
C = linop.FiniteDifference(input_shape=x_gt.shape, append=0)
solver = ADMM(
f=f,
g_list=[g_iso],
C_list=[C],
rho_list=[1e1],
x0=y,
maxiter=100,
subproblem_solver=LinearSubproblemSolver(cg_kwargs={"tol": 1e-3, "maxiter": 20}),
itstat_options={"display": True, "period": 10},
)
print(f"Solving on {device_info()}\n")
solver.solve()
x_iso = solver.x
print()
Solving on GPU (NVIDIA GeForce RTX 2080 Ti)
Iter Time Objective Prml Rsdl Dual Rsdl CG It CG Res
-----------------------------------------------------------------
0 2.91e+00 1.086e+05 1.130e+02 7.375e+02 0 0.000e+00
10 3.89e+00 3.911e+04 1.613e+01 4.388e+02 12 8.974e-04
20 4.24e+00 2.333e+04 9.468e+00 1.396e+02 15 8.943e-04
30 4.52e+00 2.228e+04 2.691e+00 2.984e+01 11 9.646e-04
40 4.74e+00 2.225e+04 1.089e+00 6.712e+00 7 9.354e-04
50 4.89e+00 2.226e+04 6.779e-01 2.817e+00 6 9.496e-04
60 5.02e+00 2.226e+04 4.808e-01 1.298e+00 3 8.914e-04
70 5.12e+00 2.227e+04 3.566e-01 8.595e-01 2 8.337e-04
80 5.21e+00 2.227e+04 2.788e-01 6.620e-01 2 9.770e-04
90 5.31e+00 2.227e+04 2.407e-01 4.794e-01 5 8.334e-04
99 5.38e+00 2.227e+04 1.948e-01 1.765e-01 1 9.823e-04
Denoise with anisotropic total variation for comparison.
[5]:
# Tune the weight to give the same data fidelity as the isotropic case.
λ_aniso = 1.2e0
g_aniso = λ_aniso * functional.L1Norm()
solver = ADMM(
f=f,
g_list=[g_aniso],
C_list=[C],
rho_list=[1e1],
x0=y,
maxiter=100,
subproblem_solver=LinearSubproblemSolver(cg_kwargs={"tol": 1e-3, "maxiter": 20}),
itstat_options={"display": True, "period": 10},
)
solver.solve()
x_aniso = solver.x
print()
Iter Time Objective Prml Rsdl Dual Rsdl CG It CG Res
-----------------------------------------------------------------
0 3.79e-01 1.165e+05 1.330e+02 8.477e+02 0 0.000e+00
10 7.20e-01 3.707e+04 2.212e+01 4.390e+02 13 9.291e-04
20 1.01e+00 2.302e+04 9.601e+00 1.202e+02 15 8.581e-04
30 1.27e+00 2.226e+04 2.706e+00 2.652e+01 11 8.191e-04
40 1.46e+00 2.224e+04 1.132e+00 7.486e+00 7 9.416e-04
50 1.59e+00 2.224e+04 6.684e-01 3.664e+00 3 9.745e-04
60 1.71e+00 2.224e+04 4.317e-01 2.308e+00 2 8.436e-04
70 1.85e+00 2.225e+04 3.064e-01 1.535e+00 2 6.615e-04
80 1.93e+00 2.225e+04 2.309e-01 1.122e+00 2 7.092e-04
90 2.04e+00 2.225e+04 1.887e-01 7.174e-01 1 8.656e-04
99 2.12e+00 2.225e+04 1.652e-01 3.941e-01 1 9.124e-04
Compute and print the data fidelity.
[6]:
for x, name in zip((x_iso, x_aniso), ("Isotropic", "Anisotropic")):
df = f(x)
print(f"Data fidelity for {name} TV was {df:.2e}")
Data fidelity for Isotropic TV was 1.93e+04
Data fidelity for Anisotropic TV was 1.92e+04
Plot results.
[7]:
plt_args = dict(norm=plot.matplotlib.colors.Normalize(vmin=0, vmax=1.5))
fig, ax = plot.subplots(nrows=2, ncols=2, sharex=True, sharey=True, figsize=(11, 10))
plot.imview(x_gt, title="Ground truth", fig=fig, ax=ax[0, 0], **plt_args)
plot.imview(y, title="Noisy version", fig=fig, ax=ax[0, 1], **plt_args)
plot.imview(x_iso, title="Isotropic TV denoising", fig=fig, ax=ax[1, 0], **plt_args)
plot.imview(x_aniso, title="Anisotropic TV denoising", fig=fig, ax=ax[1, 1], **plt_args)
fig.subplots_adjust(left=0.1, right=0.99, top=0.95, bottom=0.05, wspace=0.2, hspace=0.01)
fig.colorbar(
ax[0, 0].get_images()[0], ax=ax, location="right", shrink=0.9, pad=0.05, label="Arbitrary Units"
)
fig.suptitle("Denoising comparison")
fig.show()
# zoomed version
fig, ax = plot.subplots(nrows=2, ncols=2, sharex=True, sharey=True, figsize=(11, 10))
plot.imview(x_gt, title="Ground truth", fig=fig, ax=ax[0, 0], **plt_args)
plot.imview(y, title="Noisy version", fig=fig, ax=ax[0, 1], **plt_args)
plot.imview(x_iso, title="Isotropic TV denoising", fig=fig, ax=ax[1, 0], **plt_args)
plot.imview(x_aniso, title="Anisotropic TV denoising", fig=fig, ax=ax[1, 1], **plt_args)
ax[0, 0].set_xlim(N // 4, N // 4 + N // 2)
ax[0, 0].set_ylim(N // 4, N // 4 + N // 2)
fig.subplots_adjust(left=0.1, right=0.99, top=0.95, bottom=0.05, wspace=0.2, hspace=0.01)
fig.colorbar(
ax[0, 0].get_images()[0], ax=ax, location="right", shrink=0.9, pad=0.05, label="Arbitrary Units"
)
fig.suptitle("Denoising comparison (zoomed)")
fig.show()